CS8395 Special Topics in Computational Biology
🧭 Description
This graduate seminar examines how computational methods are designed, assessed, and advanced to tackle modern biological questions, with emphasis on applying machine learning and AI to model, integrate, and interpret single-cell and spatial transcriptomic data. In particular the course explores how choices in data representation, model specification, and algorithm design shape what can be inferred from such datasets.
Through discussions and student presentations of recent work from both conferences (e.g., ISMB, RECOMB, NeurIPS, MLCB etc.) and journals (Nature Methods, Nature Biotechnology etc.), students will learn how to critically evaluate existing models, identify the underlying assumptions and limitations, and develop their own ideas. The course also includes hands-on mini-projects in which students will design, implement, and assess a computational method inspired by the papers discussed in class.
💡 Beyond running analyses, we focus on how to think like a method developer — turning ideas into models that connect computation and biology.
Logistics
- Course Code: CS8395-04
- Term: Spring 2026
- Class Times: Tuesdays & Thursdays, 02:45 PM - 04:00 PM
- Location: Olin Hall 131
🧬 Computational biology in a Nutshell
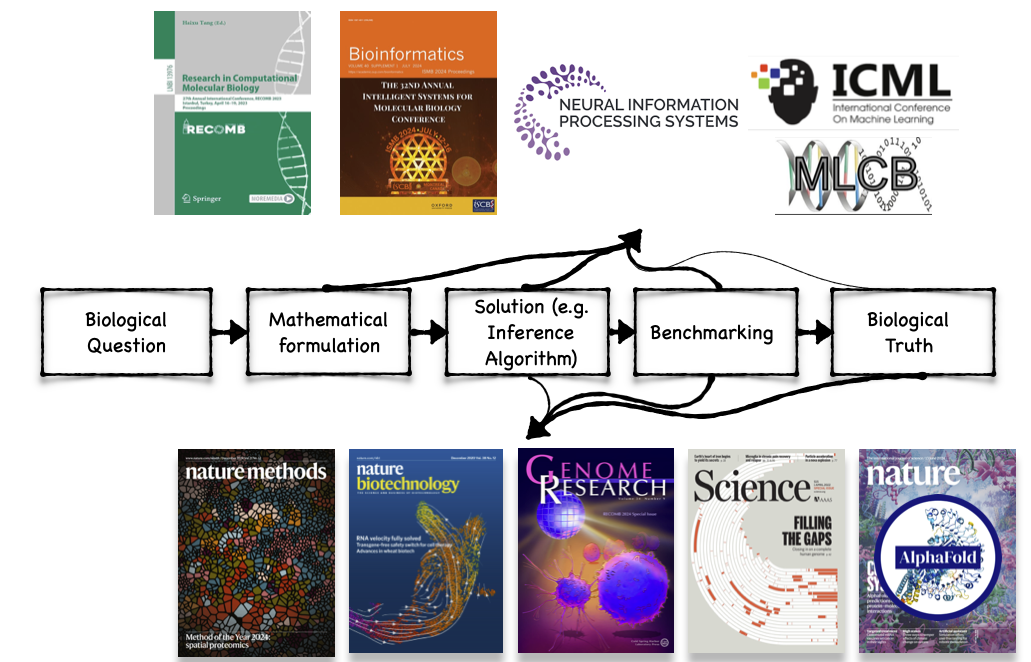
🎯 Learning Goals
By the end of this course, students will be able to:
- Define computational biology and its scope across computer science and life sciences
- Understand major biological data types — single-cell, spatial, proteomic, genomic, and multi-omics
- Formulate computational methods to address biological problems
- Evaluate and reconstruct mathematical models from research literature
- Appreciate the role of simulation and benchmarking in method development
- Translate mathematical models into efficient, reproducible implementations
🗓️ Lecture Outline
| Lecture/Date | Theme / Discussion Paper(s) | Computational Topics | Hands-on Notebook |
|---|---|---|---|
| L1/ Tue, Jan 06, 2026 | What is computational biology — Way et al., Eraslan et al.; How to read a paper | ||
| L2/ Thu, Jan 08, 2026 | In Class Project Discussion | How do we present a paper effectively? What not to do? | TBD |
| L3/ Tue, Jan 13, 2026 | Representation of data — Scanpy | Hierarchical Data Format | sc_rana_data.ipynb spatial_data.ipynb |
| L4/ Thu, Jan 15, 2026 | Dimensionality Reduction — Sun et al.;Yin et. al. | PCA, SVD, tSNE, UMAP, unsupervised/supervised clustering | low_dim_embeddings.ipynb benchmark_clustering.ipynb |
| L5/ Tue, Jan 20, 2026 | Feature Engineering for Omics - Zappia et al.;Yang et al. | Variance modeling | hvg_selection.ipynb |
| L6/ Thu, Jan 22, 2026 | Representation Learning for Omics - Eraslan et al. Benzio et al. | Autoencoders, denoising AEs, latent representations, disentanglement, nonlinear manifolds. | representation_learning.ipynb |
| L7/ Tue, Jan 27, 2026 | Clustering Algorithms — Xie et al. 2016, JMLR; Kiselev et al. 2017, Nature Meth | Discrete structure in latent space | cluster_stability.ipynb |
| L8/ Thu, Jan 29, 2026 | Trajectory & Pseudotime - Haghverdi et al. 2016, Nature Meth Qui et al. 2022, Nature Genetics | Continuous structure in latent space | pseudotime_demo.ipynb |
| L9/ Tue, Feb 03, 2026 | Initial discussion about Projects and group allocation | TBD | |
| L10/ Thu, Feb 05, 2026 | Spatial Transcriptomics — Squidpy, Palla et al. 2022, Nature Methods | Spatial data structures, spot deconvolution | TBD |
| L11/ Tue, Feb 10, 2026 | Probabilistic spatial deconvolution — DestVI, Lopez et al. 2022, Nature Biotech; RCTD, Cable et al. 2019, Nature Biotech | NMF/PNMF, VAEs for spatial data | TBD |
| L12/ Thu, Feb 12, 2026 | GNN foundation - GCN, Kipf & Welling 2016, ICLR; GraphSAGE, Hamilton et al. 2017, NeurIPS GAT, Veličković et al. 2018, ICLR | Graph representations, GNNs, | gnn_basics.ipynb |
| L13/ Tue, Feb 17, 2026 | GNN/GCN for spatial transcriptomics data - GraphST; STAGATE | Graph representations, GNNs, spatial autocorrelation | TBD |
| L14/ Thu, Feb 19, 2026 | Bridging Histology and Spatial Transcriptomics — Tangram SpaGCN iStar | histology.ipynb | |
| L15/ Tue, Feb 24, 2026 | Spatial Statistics — spatialDE; SPARK DESpace | Detecting spatially variable genes | spatial_cov.ipynb |
| L16/ Thu, Feb 26, 2026 | Segmentation I — CellPose; CellVIT | Cell Segmentation basics | spatial_cov.ipynb |
| L17/ Tue, Mar 03, 2026 | Segmentation II — Baysor; | Bayesian view of cell segmentation | spatial_cov.ipynb |
| L18/ Thu, Mar 05, 2026 | Mid semester project presentations I | TBD | |
| L19/ Tue, Mar 10, 2026 | Mid semester project presentations II | TBD | |
| Thu, Mar 12, 2026 | No class — Spring Break | ||
| Tue, Mar 17, 2026 | No class — Spring Break | ||
| L20 Tue, Mar 19, 2026 | Multi-omics integration - - Papers TBD | TBD | |
| L21 Tue, Mar 24, 2026 | Simulating multi-omics data - - Papers TBD | TBD | |
| L22 Tue, Mar 31, 2026 | Benchmarking - - Papers TBD | TBD | |
| L23 Tue, Apr 02, 2026 | Concept Recap (Theme - Model Development) | TBD | |
| L24 Tue, Apr 07, 2026 | Concept Recap (Theme - Biological Significance) | TBD | |
| L25 Tue, Apr 09, 2026 | Final Group Presentations I | TBD | |
| L26 Tue, Apr 14, 2026 | Final Group Presentations II | TBD | |
| L27 Tue, Apr 16, 2026 | Peer Review Discussion - | TBD | |
| L28 Tue, Apr 21, 2026 | Project wrap-up discussions - | TBD |
🧩 Core Topics (Summary)
- Foundations
- What is computational biology?
- Designing methods vs running tools
- Data Representation
- Single-cell, spatial, and multi-omics data
- Structuring biological datasets
- Exploratory Analysis
- Visualization, dimensionality reduction, clustering
- Feature selection in multi-omics contexts
- Problem Formulation
- Translating biological questions into computational problems
- Case studies from recent literature
- Modeling & Software
- Statistical, probabilistic, and ML frameworks
- Coding (Python, R, C++, Rust), efficiency, and reproducibility
- Simulation & Benchmarking
- Role of simulations and benchmark datasets
- Designing fair, reproducible evaluations
🏗️ Course Structure
Lectures/Discussions: Weekly topics introduced through short lectures and literature discussions (back-to-back or two days of the same week).
Readings: Research papers, book chapters, and review articles.
Assignments: Focused problem sets and small coding projects.
Final Project: Students will select a computational biology problem, survey existing literature, and either (1) reconstruct/improve an existing method or (2) propose/implement a novel approach.
👥 Evaluation (Groups)
| Component | Weight |
|---|---|
| Paper Presentation | 20% |
| - Presentation | |
| - Critical Evaluation | |
| Initial Proposal Presentation | 15% |
| Midterm Project Report | 15% |
| Midterm Project Presentation | 15% |
| Final Project Report | 15% |
| Final Project Presentation | 15% |
| Peer Review | 20% |
📚 Suggested References
- Compeau & Pevzner, BIOINFORMATICS ALGORITHMS - An Active Learning Approach
- John Tukey, Exploratory Data Analysis
- Pevzner & Shamir, Computational and Systems Biology
- Blum, Hopcroft & Kannan, Foundations of Data Science
- Single-cell Best Practices
- Selected research papers (to be assigned weekly)